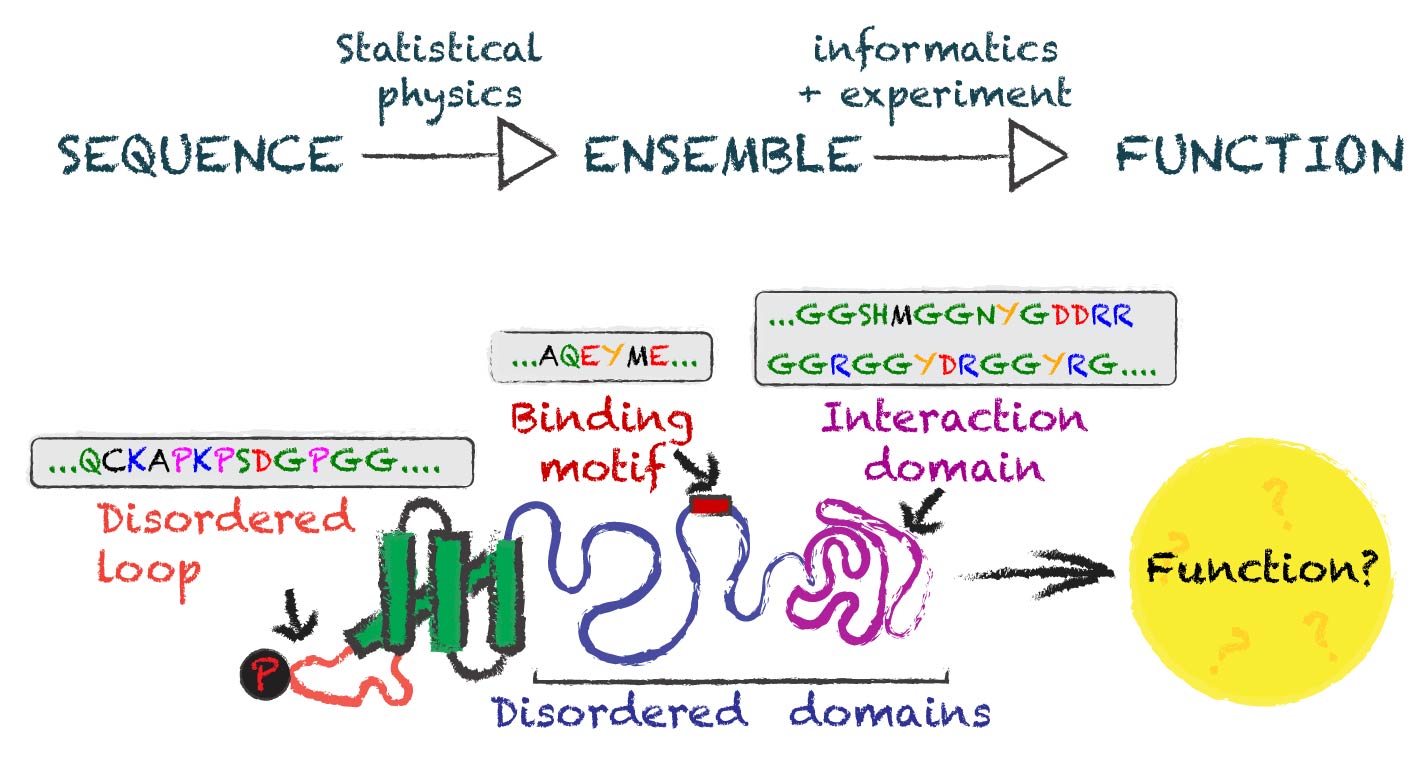
We study how function is encoded BY disordered protein REGIONS
We do this in the Department of Biochemistry and Molecular Biophysics at Washington University School of Medicine using a combination of physics-based simulations, computational biology, and experimental approaches.
TL/DR: A large fraction of proteins and protein regions are classified as "intrinsically disordered". These regions have historically been hard to study, but play key roles in a wide variety of cellular functions. Furthermore, these regions are strongly implicated in many diseases. In the Holehouse lab, we integrate a range of computational approaches (simulations, bioinformatics, systems biology) with experimental data to uncover how intrinsically disordered regions mediate cellular function, with a particular interest in biological phase separation.
Scientific Overview
We often think of proteins as tiny well-defined machines that mediate biological function in a way that is inherently linked to their 3D structure. However, many protein regions are referred to as ‘intrinsically disordered’ - regions that don’t fold into a well-defined 3D shape but instead exist in an ensemble of conformations.
These intrinsically disordered regions (IDRs) play key roles in a wide variety of cellular functions, including gene expression, signal transduction, and the stress response. They are also frequently mutated in diseases, from neurodegenerative conditions to cancer. Despite their cellular importance and clinical significance, we lack effective generalizable ways to predict and understand function from sequence.
Our lab is focused on combining molecular biophysics (computational and experimental), deep learning, and quantitative cell biology to uncover the general principles that underlie how function is encoded into disordered proteins.
Areas of interest
We are particularly interested in the following areas:
Evolution and conservation of disordered regions.
Determinants of specificity in IDR-mediated interactions (and chemical specificity in IDRs and folded domains).
Rational design of IDRs for biotechnological applications.
IDRs in transcriptional regulation.
Form and function of biomolecular condensates
Approaches
We use a wide range of experimental and computational approaches, including but not limited to:
Molecular simulations across a range of resolutions (all-atom, coarse-grained, polymer models etc.)
Deep learning (discriminative and generative)
Physics-based informatics
Yeast molecular biology
In vitro biochemistry and biophysics
LAB ETHOS
A major goal of the lab is to create a safe and supportive environment for lab members from all walks of life. Our combined experiences, perspectives, and expertise are a major asset that can only be realized by creating a professional environment that promotes and supports the success of all scientists. This support extends beyond our physical lab space to all members of our broader scientific community, and all trainees should feel welcome to contact Alex or other lab members for support and advice.
Funding Acknowledgement
The Holehouse lab has been lucky enough to receive generous support from several funding sources. We gratefully acknowledge these sources, and in particular, the American tax payer.
Active funding (9)
NIH (NHLBI) (2025 – 2029) [1R01HL179124]
Project title: Impact of sequence changes on key intrinsically disordered regions of cardiac troponin
Role: Co-Principal Investigator (MPI grant) with Michael Greenberg and Andrea Soranno
Link: NIH RePORTER
NSF (MCB) (2024 – 2029) [2338129] (NSF CAREER Award)
Project title: CAREER: Evolutionary Principles of Intrinsically Disordered Proteins
Role: Principal Investigator
Link: NSF website
Hope Center for Neurological Disorders (2024 – 2026)
Project title: Targeting a transcriptional co-repressor to prevent photoreceptor degeneration
Role: Co-Principal Investigator (with Joe Corbo)
NIH (NCI) (2023 - 2028) [DP2-CA290639] (NIH new Innovator Award)
Project title: Uncovering the Regulatory Logic of Gene Expression Encoded by Disordered Regions
Role: Principal Investigator
Link: NIH RePORTER
HFSP (2022 – 2025) [RGP0015/2022]
Project title: Molecular Determinants of Evolutionary Conservation in Disordered Protein Regions
Role: Principal Investigator (with Hyun Kate Lee and Dolf Weijers)
NSF (DBI) (2022 – 2027) [2213983]
Project title: BII: Life Without Water: Protecting Macromolecules, Cells, and Organisms During Desiccation and Rehydration Across Kingdoms of Life
Role: Co-Principal Investigator
Link: NSF website, WALII website
NSF (MCB) (2021 – 2025) [2128068]
Project title: IntBIO: Collaborative Research: Functional Synergy Between Disordered Proteins and their Environment in Desiccation Protection
Role: Principal Investigator (with Shahar Sukenik and Thomas Boothby)
Link: NSF website
NIH (NIAID) (2022 – 2027) [R01AI163142]
Project title: A Multipronged Investigation of SARS-CoV-2 Genome Packaging (with Andrea Soranno and Kathleen Hall)
Role: Co-Investigator
Link: NIH RePORTER
NIH (NIGMS) (2021 - 2025) [R01GM142164]
Project title: Intrinsic Disorder and Agonist Bias in EGF Receptor Signaling (with Linda Pike)
Role: Co-Investigator
Link: NIH RePORTER
Expired funding
Longer Life Foundation (2020 – 2023)
Predicting the Functional Impact of Genetic Variation Within Intrinsically Disordered Protein Regions
Dewpoint Therapeutics Sponsored Research Agreement (2020-2022)